International Research Resource Center in Biomolecular Mass Spectrometry and Proteomics
Current Research Portfolio
A major theme of research within the Resource is development of strategies and technology for studies involving the global detection, site assignments and dynamics of protein posttranslational modifications(PTMs). Most modifications are currently studied in isolation, but it is well known that there may be instances of cross-talk between different PTMs[1, 2, 3, 4, 6], so by studying only one modification at a time, the full regulatory process of cells cannot be revealed. In addition, most analysis protocols involve characterization of modifications after digestion of proteins into small peptides. This approach loses connectivity between multiple modifications on the same protein species. Thus we pursue development of high mass accuracy and resolution strategies to analyze peptides that are much larger than those obtained from tryptic digestions[7]
.
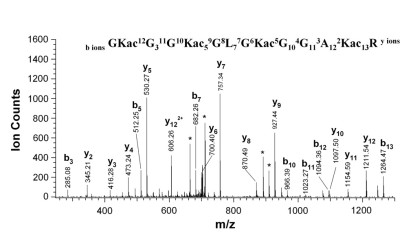
Spectrum of acylated histone H4 peptide [8]
The biological paradigm where understanding the relationship between multiple modifications is best recognized as being important is the epigenetic regulation of gene expression through the modification of histones [3, 7, 9]. As the sequence of the histones bound to all DNA is identical, regulation is achieved through dynamic modulation of combinations of either activating or repressing post-translational marks. These combinations of co-occuring modifications have been referred to as a histone code, and deconvolving these regulatory messages ideally requires strategies that can characterize intact histone proteins. The Resource is developing strategies employing electron capture dissociation (ECD) and electron transfer dissociation (ETD) to fragment intact proteins on a chromatographic time-scale[7]. This work involves development of improved chromatographic separation strategies, optimization of the latest mass spectrometric instrumentation, and production of bioinformatic tools that can rapidly and efficiently summarize the acquired data[10, 11].
Most recently we have developed a large scale multiplex pipeline for quantitation of selected phospho-peptides (also other PTMs) using new ultra long monolithic liquid capillary chromatography and parallel reaction monitoring mass spectrometry (PRM). We have shown that this is highly accurate general methodology for quantitation of some thousand selected site-specific PTMs per chromatographic run to enable studies of the dynamics of complex signaling networks [[12]. This methodology is ideal for gaining new insight into re-wiring studies not amenable to immunoaffinity methods.
The Resource has major efforts in developing strategies for the analysis of O–GlcNAcylation[13, 14] and phosphorylation[15, 16, 17]. These modifications interplay with each other in the regulation of many biological processes [6,18, 19, 20, 21], and O–GlcNAcylation has been implicated in diseases including diabetes, cancer and Alzheimer[22], so there is a major effort within the Resource to characterize cross-talk between these two types of modification. This requires development of analysis approaches that allow thorough study of both modification types from the same sample through sequential enrichment of each modification type[23]. As noted above the use of enzymes that produce large fragments is also being employed to increase the frequency of multiple modifications being present on a single peptide, to confirm their co-existence on the same protein species. This work is made more comprehensive by the availability of complementary fragmentation approaches of CID and ETD on the same instrument.
Assigning a biological role to a post-translational modification can be challenging when there are large numbers of proteins in a sample and multiple pathways are being activated. Hence, the Resource is helping develop a number of strategies that allow more targeted understanding of modifications. These include methods that allow identification of specific targets of kinase inhibitors[25], identification of phosphorylation targets of specific kinases[26, 27,28,29,30, 31, 32, 33], or identification of post-translational modifications that are regulated by activation of a specific receptor tyrosine kinase[34]. There are also projects investigating post-translational proteolytic processing to form new protein N–termini[35], which have major implications in processes such as apoptosis and inflammation, necrosis, and may have application in biomarker discovery. The human genome encodes more proteolytic enzymes than protein kinases, so many important biological events are expected to be revealed with this effort.
In addition to the work in proteomics and epigenetics above, we have focused significant effort on other studies concerning the architecture of protein complexes and machines for which angstrom resolution structural information has not yet been tractable. We determine high quality distance constraints from chemical cross-linking of the physiologically active complexes or machines, most recently in studies with the Kornberg group in contributing some 1600 obtained from mass spectral sequencing to establish the structure of the complete Mediator-RNA polymerase pre-initiation complex (52-subunits) and the regulation of transcription [36]. In addition, we have developed a new epsilon-amino lysine-lysine cross-linking strategy based on chemical reductive amination that provides comprehensive sequence and cross-link site assignments using electron transfer dissociation (ETD) [37]. This information provides accurate distance constraints that complement cryoEM and computer modeling efforts. In parallel software algorithms and scoring strategies have been developed that greatly facilitates the assignment of cross-linked peptides in general. Very recently we have initiated a thrust into development and application of methodology to measure non-covalent protein complexes directly in the gas phase using a newly acquired high mass Orbitrap Exactive instrument (m/z < 22,000) [38]. This effort will complement our long-standing work on chemical cross-linking of protein complexes and machines.
Many of these approaches are reliant on obtaining quantitative measurements and the Resource has been involved in development and application of a number of isotopic labeling approaches for quantification[16, 41, 42]. Further development of the neucode metabolic labeling strategy will enable whole animal protein quantitation[ [43]. A major present frustration of several of these strategies is the incomplete overlap in components quantified between different experiments, so the Resource is developing targeted approaches that allow sensitive quantification of the same components in each comparative experiment.[10]
Collaborations
The resource has over fifty active collaborative projects. Some of these involve application of existing technology to important biological and biomedical questions. Others are technically more challenging and require the development of new techniques, reagents or software in order to solve pressing problems. Several of these are successful long-term collaborative efforts with high-profile researchers such as Davide Ruggero, Frank McCormick, Michael Fainzilber, Harley Kornblum, Lennart Mucke, Jeff Twiss, Matt Rasband, Kevan Shokat, Elior Peles and Jack Taunton.
Protein Prospector and Other Software
For all these research projects a key component is the availability of robust and powerful bioinformatics software to manage, interpret and summarize results. The Resource develops arguably the most powerful and diverse suite of proteomic analysis tools available in Protein Prospector. Peptide and protein identification from tandem mass spectrometry data is the cornerstone of most current proteomic research and Protein Prospector has been demonstrated to be one of the most sensitive and effective search engines for interpretation of data produced from a wide range of different instruments[44]. The recent widespread emergence of ECD and ETD fragmentation approaches for proteomic research presents new challenges for searching software, but Protein Prospector has been shown to be one of the most effective at analysis of this type of data[45].
The ability to compare or combine results from multiple datasets is an important part of proteomic research, but this functionality is not available in most search engines. However, Protein Prospector uses a database structure for storing previous search results that allows comparison or merging of unlimited numbers of different data analyses[46]. This has particular application in studies where CID and ETD analysis of the same sample may have been performed and a combined results summary is desired.
Relative quantification of components can be determined using isotopic labeling or label-free strategies. Protein Prospector supports both these measurement strategies, and is the only freely available software that combines database searching and stable isotope quantification analysis in one package.
Chemical cross-linking strategies are powerful for characterizing protein structure, binding partners and interaction surfaces. However, analysis of this type of data is complicated. The Resource is developing improved searching strategies within Protein Prospector that allow identification of cross-linked peptides in the background of complex peptide mixtures[47].
All these features of Protein Prospector are freely available through a public website maintained by the Resource. This website is used for over a million searches every year, demonstrating its importance to the research community. Researchers are also able to contact us through the website to request new features, and these suggestions are evaluated and those seen as being of wide use to other researchers are implemented.
Training and Dissemination
Training in proteomic analysis strategies and dissemination of technologies developed within the facility are significant parts of the work of the Resource. In support of this aspect of the Resource's mission, the facility is continuously working with and mentoring postdoctoral scholars and graduate students. Currently the facility's research staff includes six postdoctoral scholars and three graduate students. Additionally, one graduate-level course is taught by the Resource (Chem204). It is open to other researchers, and a number of post-doctoral scholars and PIs from both within UCSF and from other local research institutions attend. The Resource also hosts a biennial International Symposium on Mass Spectrometry in the Health and Life Sciences, which attracts world-class researchers from both mass spectrometry and biological backgrounds and is a major avenue for fertilization of cross-disciplinary research.
The Resource publishes extensively in peer-reviewed journals (available on the website) and also presents its research at a range of international conferences and forums. Through this website it also makes available protocols (In-Gel Digestion, Phosphorylated Peptide Enrichment Using TiO2, and Lectin Weak Affinity Chromatography) and software tools (ProteinProspector and available software) used in the Resource.
References
[1] Cohen P, The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci, 2000. 25(12): 596-601. [Pubmed]
[2] Benayoun BA, Veitia RA, A post-translational modification code for transcription factors: sorting through a sea of signals. Trends Cell Biol, 2009. 19(5): 189-97. [Pubmed]
[3] Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ, How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol, 2007. 14(11): 1025-40. [Pubmed]
[4] Yang XJ, Multisite protein modification and intramolecular signaling. Oncogene, 2005. 24(10): 1653-62. [Pubmed]
[5] Hunter T, The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell, 2007. 28(5): 730-8. [Pubmed]
[6] Kaasik K, Kivimäe S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptáček LJ, Fu YH., Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab., 2013. 17(2):291-302. [Pubmed] [7] Eliuk SM, Maltby D, Panning B, Burlingame AL., High resolution electron transfer dissociation studies of unfractionated intact histones from murine embryonic stem cells using on-line capillary LC separation: determination of abundant histone isoforms and post-translational modifications. Mol Cell, 2010. 9(5):824-37. [Pubmed] [8] Casati P, Campi M, Chu F, Suzuki N, Maltby D, Guan S, Burlingame AL and Walbot V., Histone Acetylation and Chromatin Remodeling Are Required for UV-B–Dependent Transcriptional Activation of Regulated Genes in Maize. The Plant Cell, 2008. 20: 827-842.. [Pubmed][9] Strahl BD, Allis CD, The language of covalent histone modifications. Nature, 2000. 403(6765): 41-5. [Pubmed]
[10] Guan S, Burlingame AL., High mass selectivity for top-down proteomics by application of SWIFT technology. J Am Soc Mass Spectrom., 2010. 21(3):455-9. [Pubmed]
[11] Guan S, Burlingame AL, Data processing algorithms for analysis of high resolution MSMS spectra of peptides with complex patterns of posttranslational modifications. Mol Cell Proteomics, 2010. 9(5):804-10. [Pubmed]
[12] Urisman A, Levin RS, Gordan JD, Webber JT, Hernandez H, Ishihama Y, Shokat KM,Burlingame AL, An Optimized Chromatographic Strategy for Multiplexing In ParallelReaction Monitoring Mass Spectrometry: Insights from Quantitation of ActivatedKinases. Mol Cell Proteomics, 2017. 16(2):265-277. [Pubmed]
[13] Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, and Burlingame AL, O–linked N–acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics, 2006. 5(5): 923-34. [Pubmed]
[14] Chalkley RJ, Thalhammer A, Schoepfer R, and Burlingame AL, Identification of protein O–GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci U S A, 2009. 106(22): 8894-9. [Pubmed]
[15] Trinidad JC, Specht CG,Thalhammer A, Schoepfer R, and Burlingame AL, Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol Cell Proteomics, 2006. 5(5): 914-22. [Pubmed]
[16] Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, and Burlingame AL, Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics, 2008. 7(4): 684-96. [Pubmed]
[17] Trinidad JC, Thalhammer A, Specht CG, Schoepfer R, and Burlingame AL, Phosphorylation state of postsynaptic density proteins. J Neurochem, 2005. 92(6): 1306-16. [Pubmed]
[18] Wang Z, Gucek M, and Hart GW, Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O–GlcNAc. Proc Natl Acad Sci U S A, 2008. 105(37): 13793-8. [Pubmed]
[19] Wang Z, Pandey A, and Hart GW, Dynamic interplay between O–linked N–acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol Cell Proteomics, 2007. 6(8):1365-79. [Pubmed]
[20] Copeland RJ, Bullen JW, and Hart GW, Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab, 2008. 295(1):E17-28. [Pubmed]
[21] Hart GW, Housley MP, Slawson C, Cycling of O–linked beta-N–acetylglucosamine on nucleocytoplasmic proteins. Nature, 2007. 446(7139): 1017-22. [Pubmed]
[22] Dias WB and Hart GW, O–GlcNAc modification in diabetes and Alzheimer's disease. Mol Biosyst, 2007. 3(11):766-72. [Pubmed]
[23] Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL, Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012. 11(8):215-29. [Pubmed]
[24] Cohen MS, Zhang C, Shokat KM, and Taunton J, Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science, 2005. 308(5726): 1318-21. [Pubmed]
[25] Zhao Q, Ouyang X, Wan X, Gajiwala KS, Kath JC, Jones LH, Burlingame AL, Taunton J, Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes. J Am Chem Soc., 2017. 139(2):680-685. [Pubmed]
[26] Hertz NT, Wang BT, Allen JJ, Zhang C, Dar AC, Burlingame AL, Shokat KM, Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr Protoc Chem Biol., 2010 2(1): 15-36. [Pubmed]
[27] Shah K, Liu Y, Deirmengian C, and Shokat KM, Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci U S A, 1997. 94(8):3565-70. [Pubmed]
[28] Blethrow J, Zhang C, Shokat KM, and Weiss EL, Design and use of analog-sensitive protein kinases. Curr Protoc Mol Biol, 2004. Chapter 18: Unit 18.11. [Pubmed]
[29] Levin RS, Hertz NT, Burlingame AL, Shokat KM, Mukherjee S, Innate immunity kinase TAK1 phosphorylates Rab1 on a hotspot for posttranslational modifications by host and pathogen. Proc Natl Acad Sci U S A, 2016 . 113(33):E4776-83. [Pubmed]
[30] Ultanir SK, Yadav S, Hertz NT, Oses-Prieto JA, Claxton S, Burlingame AL, Shokat KM, Jan LY, Jan YN, MST3 kinase phosphorylates TAO1/2 to enable Myosin Va function in promoting spine synapse development. Neuron, 2014. 84: (5):968-82. [Pubmed]
[31] Sos ML, Levin RS, Gordan JD, Oses-Prieto JA, Webber JT, Salt M, Hann B, Burlingame AL, McCormick F, Bandyopadhyay S, Shokat KM, Oncogene mimicry as a mechanism of primary resistance to BRAF inhibitors. Cell Rep. 2014. 8(4):1037-48. [Pubmed]
[32] Hertz NT, Berthet A, Sos ML, Thorn KS, Burlingame AL, Nakamura K, Shokat KM, A neo-substrate that amplifies catalytic activity of parkinson's-disease-related kinase PINK1. Cell, 2013 . 154(4):737-47. [Pubmed]
[33] Hengeveld RC, Hertz NT, Vromans MJ, Zhang C, Burlingame AL, Shokat KM, Lens SM, Development of a chemical genetic approach for human aurora B kinase identifies novel substrates of the chromosomal passenger complex. Mol Cell Proteomics, 2012 11(5):47-59. [Pubmed]
[34] Raffioni S, Thomas D, Foehr ED, Thompson LM, and Bradshaw RA, Comparison of the intracellular signaling responses by three chimeric fibroblast growth factor receptors in PC12 cells. Proc Natl Acad Sci U S A, 1999. 96(13): 7178-83. [Pubmed]
[35] Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, and Wells JA, Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell, 2008. 134(5):866-76. [Pubmed]
[36] Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD, Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell, 2016. 166(6): 1411-1422.e16. [Pubmed]
[37] Trnka MJ, Burlingame AL, Topographic studies of the GroEL-GroES chaperonin complex by chemical cross-linking using diformyl ethynylbenzene: the power of high resolution electron transfer dissociation for determination of both peptide sequences and their attachment sites. Mol Cell Proteomics, 2010 9(10):2306-17. [Pubmed]
[38] Li S, Yen L, Pastor WA, Johnston JB, Du J, Shew CJ, Liu W, Ho J, Stender B, Clark AT, Burlingame AL, Daxinger L, Patel DJ, Jacobsen SE, Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc Natl Acad Sci U S A. 2016. 1113(35):E5108-16. [Pubmed]
[39] Lu J, Trnka MJ, Roh SH, Robinson PJ, Shiau C, Fujimori DG, Chiu W, Burlingame AL, Guan S, Improved Peak Detection and Deconvolution of Native Electrospray Mass Spectra from Large Protein Complexes. J Am Soc Mass Spectrom, 2015. 26(12):2141-51. [Pubmed]
[40] Guan S, Trnka MJ, Bushnell DA, Robinson PJ, Gestwicki JE, Burlingame AL, Deconvolution method for specific and nonspecific binding of ligand to multiprotein complex by native mass spectrometry. Anal Chem., 2015. 87(16):8541-6. [Pubmed]
[41] Hansen KC, Schmitt-Ulms G, Chalkley RJ, Hirsch J, Baldwin MA, and Burlingame AL, Mass Spectrometric Analysis of Protein Mixtures at Low Levels Using Cleavable 13C-Isotope-coded Affinity Tag and Multidimensional Chromatography. Mol Cell Proteomics, 2003. 2(5): 299-314. [Pubmed]
[42] Hirsch J, Hansen KC, Sapru A, Frank JA, Chalkley RJ, Fang X, Trinidad JC, Baker P, Burlingame AL, and Matthay MA, Impact of low and high tidal volumes on the rat alveolar epithelial type II cell proteome. Am J Respir Crit Care Med, 2007. 175(10): 1006-13. [Pubmed]
[43] Baughman JM, Rose CM, Kolumam G, Webster JD, Wilkerson EM, Merrill AE, Rhoads TW, Noubade R, Katavolos P, Lesch J, Stapleton DS, Rabaglia ME, Schueler KL, Asuncion R, Domeyer M, Zavala-Solorio J, Reich M, DeVoss J, Keller MP, Attie AD, Hebert AS, Westphall MS, Coon JJ, Kirkpatrick DS, Dey A, NeuCode Proteomics Reveals Bap1 Regulation of Metabolism. Cell Rep., 2016. 16(2):583-95. [Pubmed]
[44] Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, and Burlingame AL, In-depth Analysis of Tandem Mass Spectrometry Data from Disparate Instrument Types. Mol Cell Proteomics, 2008. 7(12): 2386-98. [Pubmed]
[45] Chalkley RJ, Baker PR, Lynn AJ, and Burlingame AL. Database Analysis of Electron Transfer Dissociation Mass Spectrometry Data. in ABRF 2009: Application and Optimization of Existing and Emerging Biotechnologies. 2009. Memphis, TN.
[46] Chalkley RJ, Baker PR, Huang L, Hansen KC, Allen NP, Rexach M, and Burlingame AL, Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer: II. New developments in Protein Prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol Cell Proteomics, 2005. 4(8):1194-1204. [Pubmed]
[47] Trnka MJ, Baker PR, Robinson PJ, Burlingame AL, Chalkley RJ, Matching cross-linked peptide spectra: only as good as the worse identification. Mol Cell Proteomics, 2014. 13(2):420-34. [Pubmed]